Resurrection of ancestral proteins
Investigations of the past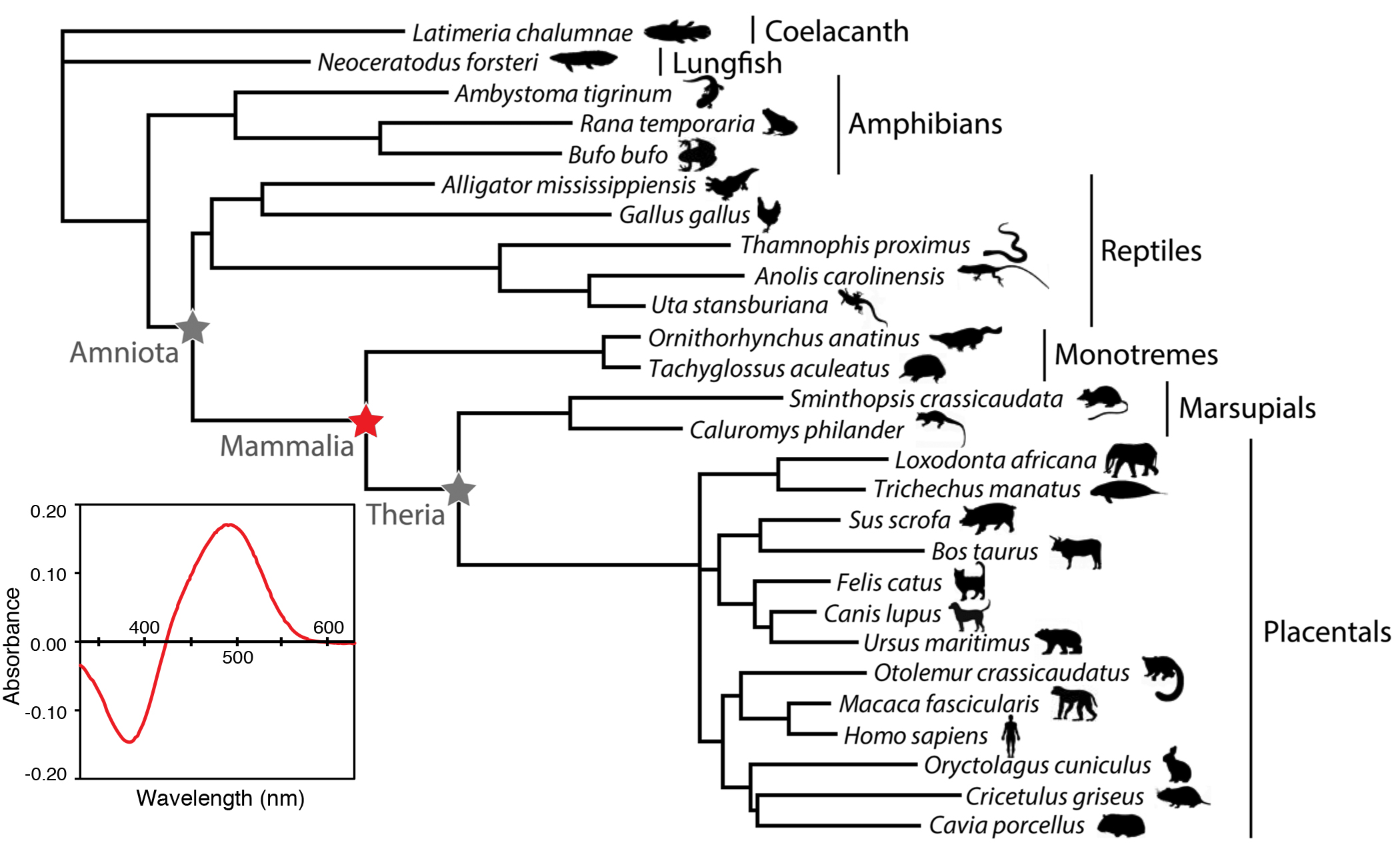 are an essential tool for understanding the present day molecular structure and function of proteins. Leveraging the rapidly expanding amounts of sequence information available in genome databases to infer the sequences of ancestral proteins, and then resurrecting them in the laboratory, is a powerful way to study the complex and intricate relationship of protein structure and function. Studies from our lab show that not only is it feasible to reconstruct ancestral sequences in the laboratory, but that this can lead to insight into the evolution of protein function not easily attainable by other methods. Moreover, these studies can also offer intriguing insights into the paleobiology of the ancient organisms in which these resurrected proteins resided. We have used these methods to study the evolution of vertebrate visual pigments, photosensitive molecules in the eye that form the first critical step in the visual transduction cascade within animal photoreceptors. This interdisciplinary approach involves using computational methods of evolutionary sequence analysis to infer ancestral sequences, synthesizing the ancestral genes, and expressing them in the laboratory. This is a longstanding research interest in our laboratory. We pioneered ancestral approaches for studying visual pigment evolution in ancestral archosaurs, and are currently applying these methods to study the origins of dim light vision in mammals, and the evolution of spectral tuning in different vertebrate groups including ongoing work in cetaceans, Neotropical fishes, and avian visual pigments, including bowerbirds and warblers. Our expertise in ancestral resurrection has led to exciting collaborative studies in other molecular systems as well. These include an evolutionary study of metabotropic glutamate receptors that has important implications for the origins of the nervous system in metazoans, as well as a study of the evolution of fluorescent proteins in corals.
are an essential tool for understanding the present day molecular structure and function of proteins. Leveraging the rapidly expanding amounts of sequence information available in genome databases to infer the sequences of ancestral proteins, and then resurrecting them in the laboratory, is a powerful way to study the complex and intricate relationship of protein structure and function. Studies from our lab show that not only is it feasible to reconstruct ancestral sequences in the laboratory, but that this can lead to insight into the evolution of protein function not easily attainable by other methods. Moreover, these studies can also offer intriguing insights into the paleobiology of the ancient organisms in which these resurrected proteins resided. We have used these methods to study the evolution of vertebrate visual pigments, photosensitive molecules in the eye that form the first critical step in the visual transduction cascade within animal photoreceptors. This interdisciplinary approach involves using computational methods of evolutionary sequence analysis to infer ancestral sequences, synthesizing the ancestral genes, and expressing them in the laboratory. This is a longstanding research interest in our laboratory. We pioneered ancestral approaches for studying visual pigment evolution in ancestral archosaurs, and are currently applying these methods to study the origins of dim light vision in mammals, and the evolution of spectral tuning in different vertebrate groups including ongoing work in cetaceans, Neotropical fishes, and avian visual pigments, including bowerbirds and warblers. Our expertise in ancestral resurrection has led to exciting collaborative studies in other molecular systems as well. These include an evolutionary study of metabotropic glutamate receptors that has important implications for the origins of the nervous system in metazoans, as well as a study of the evolution of fluorescent proteins in corals.
Engineering biological systems for the study of protein evolution and function
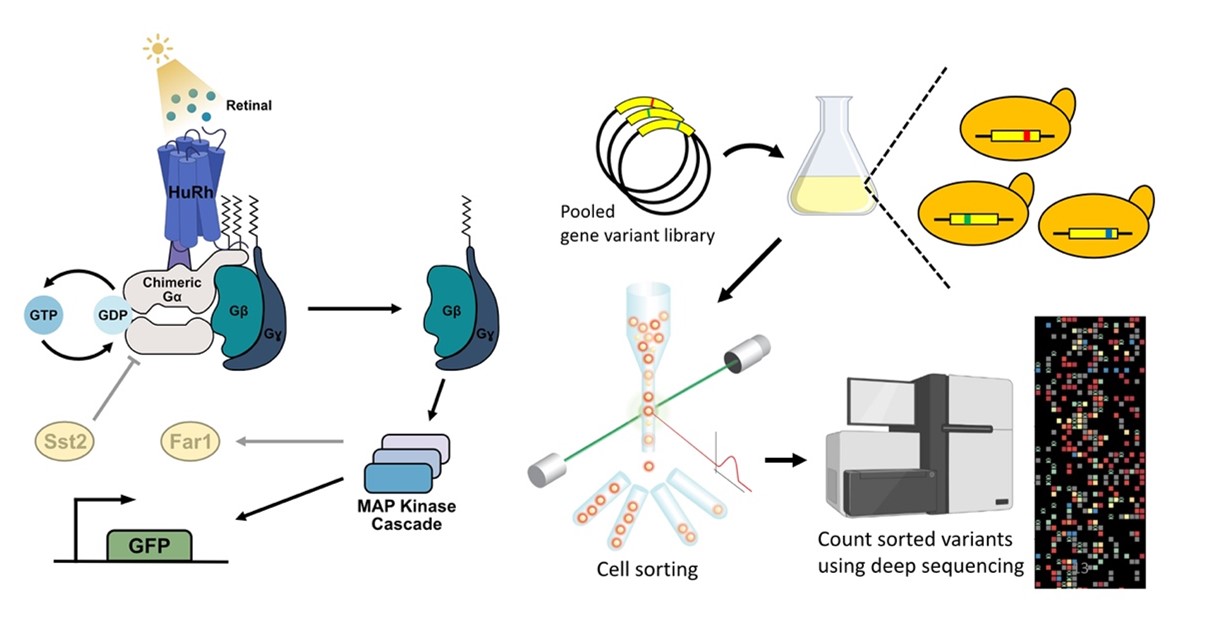 Recent breakthroughs in sequencing technology have led to the emergence of high-throughput experimental designs known as deep mutational scanning (DMS), a revolutionary approach that enables the characterization of thousands of protein variants in a single experiment. This approach has crucial implications not only for studies of human disease variants but also for evolutionary studies of protein mutational landscape, that have only recently begun to be explored. To study the mutational landscape for the visual pigment rhodopsin, we have engineered the first high-throughput assay for light-activated rhodopsin in yeast, via coupling to an endogenous G protein-coupled receptor MAP kinase pathway that results in signaling of a fluorescent reporter. We are now combining this engineered yeast system with a DMS approach that involves fluorescence-activated cell sorting and deep sequencing to examine how sequence variation translates into changes in rhodopsin function.
Recent breakthroughs in sequencing technology have led to the emergence of high-throughput experimental designs known as deep mutational scanning (DMS), a revolutionary approach that enables the characterization of thousands of protein variants in a single experiment. This approach has crucial implications not only for studies of human disease variants but also for evolutionary studies of protein mutational landscape, that have only recently begun to be explored. To study the mutational landscape for the visual pigment rhodopsin, we have engineered the first high-throughput assay for light-activated rhodopsin in yeast, via coupling to an endogenous G protein-coupled receptor MAP kinase pathway that results in signaling of a fluorescent reporter. We are now combining this engineered yeast system with a DMS approach that involves fluorescence-activated cell sorting and deep sequencing to examine how sequence variation translates into changes in rhodopsin function.
Evolutionary transformations of photoreceptors in snakes
Snakes are unique among terrestrial vertebrates in the structure and organization of their visual systems, including many unusual characteristics such as their method of accommodation and loss of ciliary muscles in the eye. Vertebrate retinas are generally composed of rod (dim-light) and cone (bright-light) photoreceptors with distinct morphologies that evolved as adaptations to nocturnal/crepuscular and diurnal light environments. Over 70 years ago, Walls proposed the ‘transmutation’ theory to explain some of the rare exceptions in which a photoreceptor type is missing, suggesting that evolutionary transitions between photoreceptor cell types. Snake visual systems are particularly fascinating for their diversity of retinal organization, which range from pure-rod retinas found in some nocturnal species, to the pure-cone retinas of many diurnal colubrid snakes. We are investigating the all-cone retina of the garter snake (Thamnophis proximus) as a model system to better understand the evolutionary fate of the rod photoreceptors in diurnal colubrid snakes. Although the ‘transmutation’ idea had been proposed decades ago, conclusive evidence supporting its validity, beyond the presence of rhodopsin sequences, had been lacking. Our investigations revealed that the rhodopsin gene sequences of the garter snake were not only functional in vitro, antibody staining demonstrated their presence in retinal cone photoreceptors, along with other components of the rod visual transduction machinery, and rod-like ultrastructural features. Future studies will address whether these features also exist in other colubrid snakes, and if there is evidence that these cone-like rods may have evolved to regain spectral sensitivity and chromatic discrimination as a result of ancestral losses of middle-wavelength cone opsins in early snake evolution.
Discerning patterns of selection and adaptation in response to visual ecology in aquatic environments
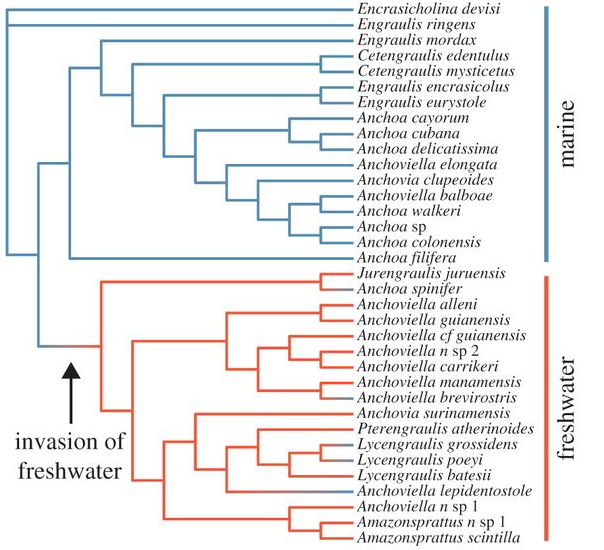 Due to their striking diversity of spectral composition, aquatic systems provide a rich source of natural experiments in which to study the effects of different light environments on the evolution of vision. We are interested in using aquatic animals to identify instances of adaptive evolution in visual proteins through investigations of natural sequence variation. Our approach incorporates computational methods to detect changes in selective constraint (dN/dS), combined with experimental assays of visual pigment function in order to empirically test hypotheses of adaptive evolution. Our goal is to better understand the molecular mechanisms underlying adaptations to aspects of visual ecology, and many of our projects involve close collaboration with field biologists. We are studying visual adaptations in several different fish systems. Paleogeographical changes in South America provide excellent opportunities for the study of visual adaptations associated with dramatic shifts in visual ecology. Incursions of marine water into large portions of South America during the Miocene are thought to be associated with the colonization of freshwater habitats by ancestrally marine species in several lineages of neotropical fishes. In an ongoing collaboration with N. Lovejoy (UTSC), we found significant evidence for high levels of positive selection in lineages of marine-derived freshwater anchovies that are associated with differences in visual ecology, along with adaptive substitutions at amino acid residues known to be associated with spectral tuning. We are currently expanding our studies to include other marine-derived freshwater lineages.
Due to their striking diversity of spectral composition, aquatic systems provide a rich source of natural experiments in which to study the effects of different light environments on the evolution of vision. We are interested in using aquatic animals to identify instances of adaptive evolution in visual proteins through investigations of natural sequence variation. Our approach incorporates computational methods to detect changes in selective constraint (dN/dS), combined with experimental assays of visual pigment function in order to empirically test hypotheses of adaptive evolution. Our goal is to better understand the molecular mechanisms underlying adaptations to aspects of visual ecology, and many of our projects involve close collaboration with field biologists. We are studying visual adaptations in several different fish systems. Paleogeographical changes in South America provide excellent opportunities for the study of visual adaptations associated with dramatic shifts in visual ecology. Incursions of marine water into large portions of South America during the Miocene are thought to be associated with the colonization of freshwater habitats by ancestrally marine species in several lineages of neotropical fishes. In an ongoing collaboration with N. Lovejoy (UTSC), we found significant evidence for high levels of positive selection in lineages of marine-derived freshwater anchovies that are associated with differences in visual ecology, along with adaptive substitutions at amino acid residues known to be associated with spectral tuning. We are currently expanding our studies to include other marine-derived freshwater lineages.
Another neotropical group, cichlids are among th e most species-rich fish groups in the Neotropics, differing from their African rift lake relatives in that their diversification took place primarily in rivers. We are investigating the molecular evolution of visual pigments in several families of Neotropical cichlids, an ongoing collaboration with H. Lopez-Fernandez (Royal Ontario Museum) and H. Rodd (U of Toronto). We found high levels of positive selection in Neotropical river cichlid rhodopsins, but interestingly, at non-overlapping subsets of amino acids sites when compared with African rift lake cichlids, suggestive of divergent selection that may target similar molecular functions. Our studies of the predatory Trinidadian pike cichlid revealed a dramatic reduction in visual pigment gene complement consistent with loss of visual sensitivity at the shorter wavelength end of the spectrum. Future work will address whether this loss is widespread among cichlids, and whether it may be associated with particular riverine environments.
e most species-rich fish groups in the Neotropics, differing from their African rift lake relatives in that their diversification took place primarily in rivers. We are investigating the molecular evolution of visual pigments in several families of Neotropical cichlids, an ongoing collaboration with H. Lopez-Fernandez (Royal Ontario Museum) and H. Rodd (U of Toronto). We found high levels of positive selection in Neotropical river cichlid rhodopsins, but interestingly, at non-overlapping subsets of amino acids sites when compared with African rift lake cichlids, suggestive of divergent selection that may target similar molecular functions. Our studies of the predatory Trinidadian pike cichlid revealed a dramatic reduction in visual pigment gene complement consistent with loss of visual sensitivity at the shorter wavelength end of the spectrum. Future work will address whether this loss is widespread among cichlids, and whether it may be associated with particular riverine environments.
Cetaceans are the only mammals that have evolved a fully aquatic lifestyle and diversified into all major aquatic ecosystems, a remarkable transition that has been characterized by extreme morphological and physiological adaptations that are not well understood at the molecular level. We are using site-directed mutagenesis combined with in vitro expression studies to investigate the spectral effects of amino acid substitutions thought to be associated with adaptations to aquatic life histories in cetaceans. We also found patterns of ecological divergence in rhodopsin evolution correlated with foraging depth, suggesting that foraging dives might be a selective regime influencing visual adaptation in cetaceans. Future work will investigate if other aspects of visual pigment function might be similarly tuned for aquatic environments in cetaceans.
Non-model systems for the study of visual pigment structure and function
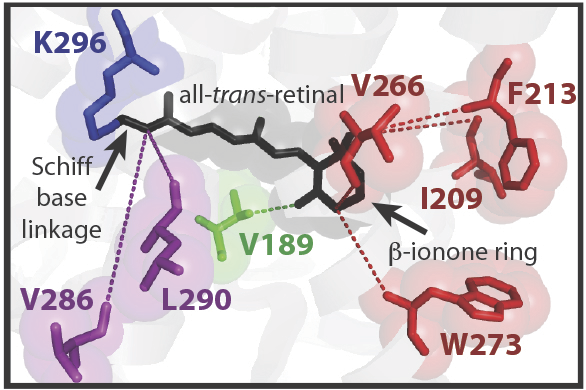 With the notable exception of spectral tuning studies, most experimental investigations of visual pigments have centered on a small number of model systems, such as bovine rhodopsin. We are interested in developing alternative model systems in order to study important aspects of visual pigment function, for aspects of function beyond spectral tuning. In order to more easily express visual pigments isolated from different animals, our lab engineered an expression vector for heterologous opsin expression in mammalian cell culture, and we have developed spectroscopic assays for visual pigment function that can be applied to non-model vertebrate pigments. We have been developing zebrafish as a model system for investigating visual pigment structure and function. As an ectothermic vertebrate, we might expect significant differences in kinetics with mammalian visual pigments. In a site-directed mutagenesis study, we were able to identify amino acid residues associated with differences in light-activated stability of rhodopsin; these differences contribute to increased rates in zebrafish rhodopsin that may be necessary for function at lower physiological temperatures. We discovered a completely novel type of visual pigment in zebrafish, Rh1-2, and characterized its expression pattern, molecular properties, as well as its evolutionary origins. We have also investigated light-activated rhodopsin kinetics in the nocturnal echidna, and in the great bowerbird, a model system for the study of sexual selection and visual behaviour.
With the notable exception of spectral tuning studies, most experimental investigations of visual pigments have centered on a small number of model systems, such as bovine rhodopsin. We are interested in developing alternative model systems in order to study important aspects of visual pigment function, for aspects of function beyond spectral tuning. In order to more easily express visual pigments isolated from different animals, our lab engineered an expression vector for heterologous opsin expression in mammalian cell culture, and we have developed spectroscopic assays for visual pigment function that can be applied to non-model vertebrate pigments. We have been developing zebrafish as a model system for investigating visual pigment structure and function. As an ectothermic vertebrate, we might expect significant differences in kinetics with mammalian visual pigments. In a site-directed mutagenesis study, we were able to identify amino acid residues associated with differences in light-activated stability of rhodopsin; these differences contribute to increased rates in zebrafish rhodopsin that may be necessary for function at lower physiological temperatures. We discovered a completely novel type of visual pigment in zebrafish, Rh1-2, and characterized its expression pattern, molecular properties, as well as its evolutionary origins. We have also investigated light-activated rhodopsin kinetics in the nocturnal echidna, and in the great bowerbird, a model system for the study of sexual selection and visual behaviour.
Functional defects in visual pigments associated with retinal disease
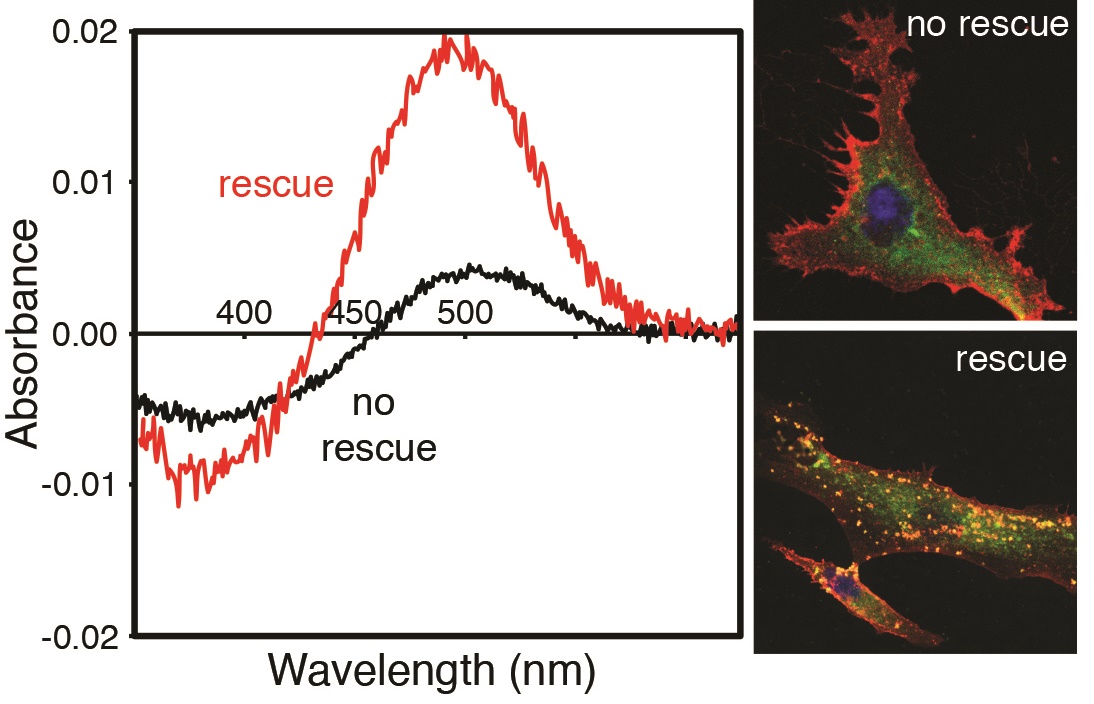 We have recently become interested in investigating disease variants in visual transduction genes. Retinitis pigmentosa (RP) is a degenerative retinal disorder often linked to mutations in rhodopsin and other elements of the visual transduction cascade. Despite advances in diagnosis and clinical characterization, molecular characterization of RP remains challenging due to the diversity of disease genes and the numerous disease-associated mutations in these genes. Rhodopsin, the visual pigment that initiates the phototransduction cascade, is the best characterized RP gene to date and accounts for 20–30% of autosomal dominant RP cases. As part of an ongoing collaboration with an ophthalmologist, E. Heon (Sick Kids Hospital), we are interested in investigating rhodopsin mutations that are associated with retinitis pigmentosa in retinal disease patients, in order to better understand aspects of rhodopsin structure and function that may be affected.
We have recently become interested in investigating disease variants in visual transduction genes. Retinitis pigmentosa (RP) is a degenerative retinal disorder often linked to mutations in rhodopsin and other elements of the visual transduction cascade. Despite advances in diagnosis and clinical characterization, molecular characterization of RP remains challenging due to the diversity of disease genes and the numerous disease-associated mutations in these genes. Rhodopsin, the visual pigment that initiates the phototransduction cascade, is the best characterized RP gene to date and accounts for 20–30% of autosomal dominant RP cases. As part of an ongoing collaboration with an ophthalmologist, E. Heon (Sick Kids Hospital), we are interested in investigating rhodopsin mutations that are associated with retinitis pigmentosa in retinal disease patients, in order to better understand aspects of rhodopsin structure and function that may be affected.
In a disease context, we are particularly interested in understanding the limits of natural variation in vertebrate visual genes so that better predictive tools for identifying disease associated mutations in humans can be developed. This line of research has led to a number of papers investigating natural variation in components of the visual transduction cascade and in the eye, and most recently, a paper investigating the relationship between natural sequence variation in visual genes and mutations associated with visual degenerative disease in humans.
Evolutionary bioinformatics/computational methods development
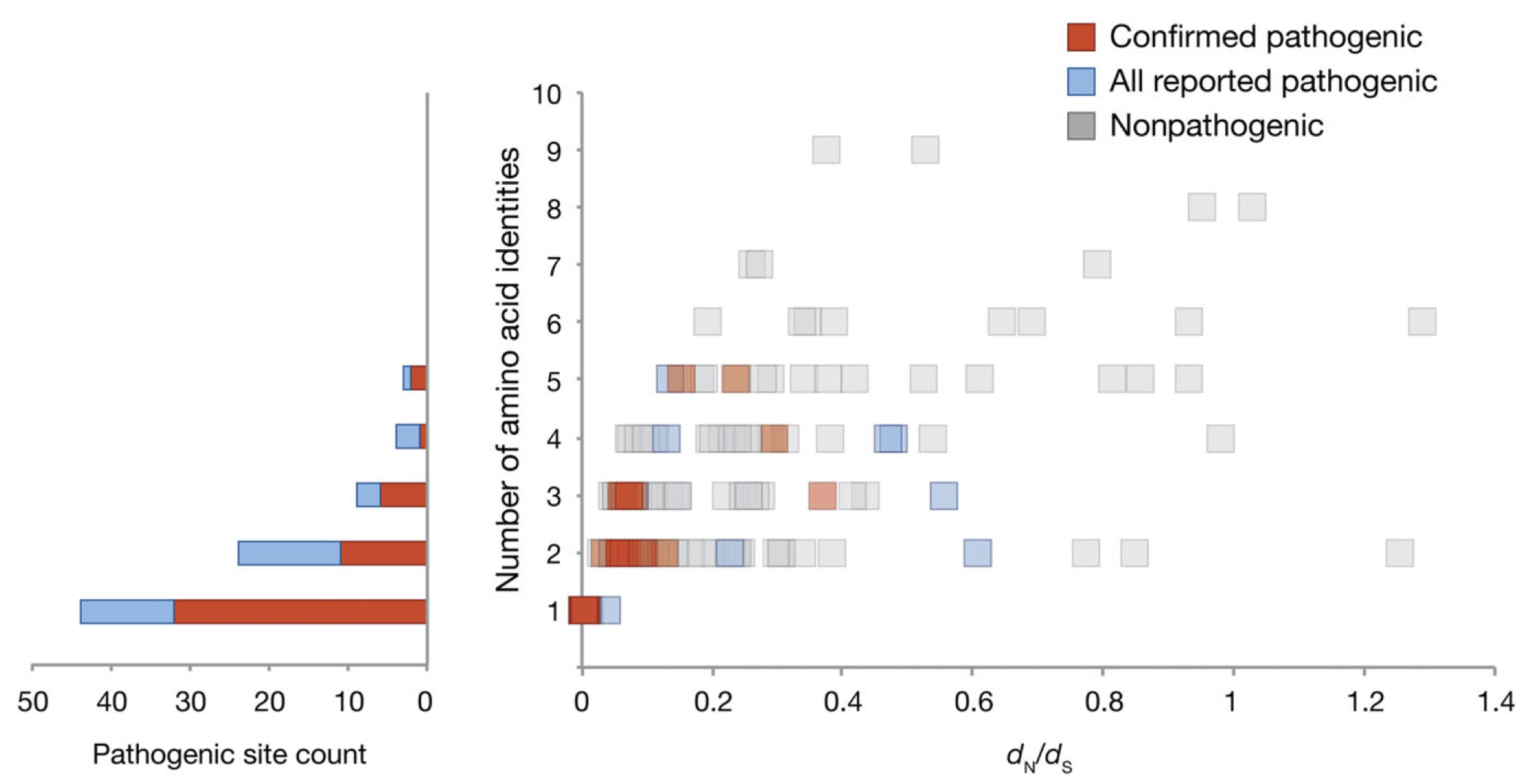 Our lab is working to refine codon-based computational methods to detect
Our lab is working to refine codon-based computational methods to detect
selective constraint in protein-coding sequences. These models rely on the estimation of a non-synonymous to synonymous substitution rate ratio, which is thought to reflect the form and strength of selection. We found that a particular class of models, the clade models, had a particularly high false positive rate in detecting the significant divergence among groups of sequences, and that we could correct for this by modifying the null model. We also provided a framework for testing for positive selection using clade models, as well as a means of testing for correlations with other variables such as ecological factors.
